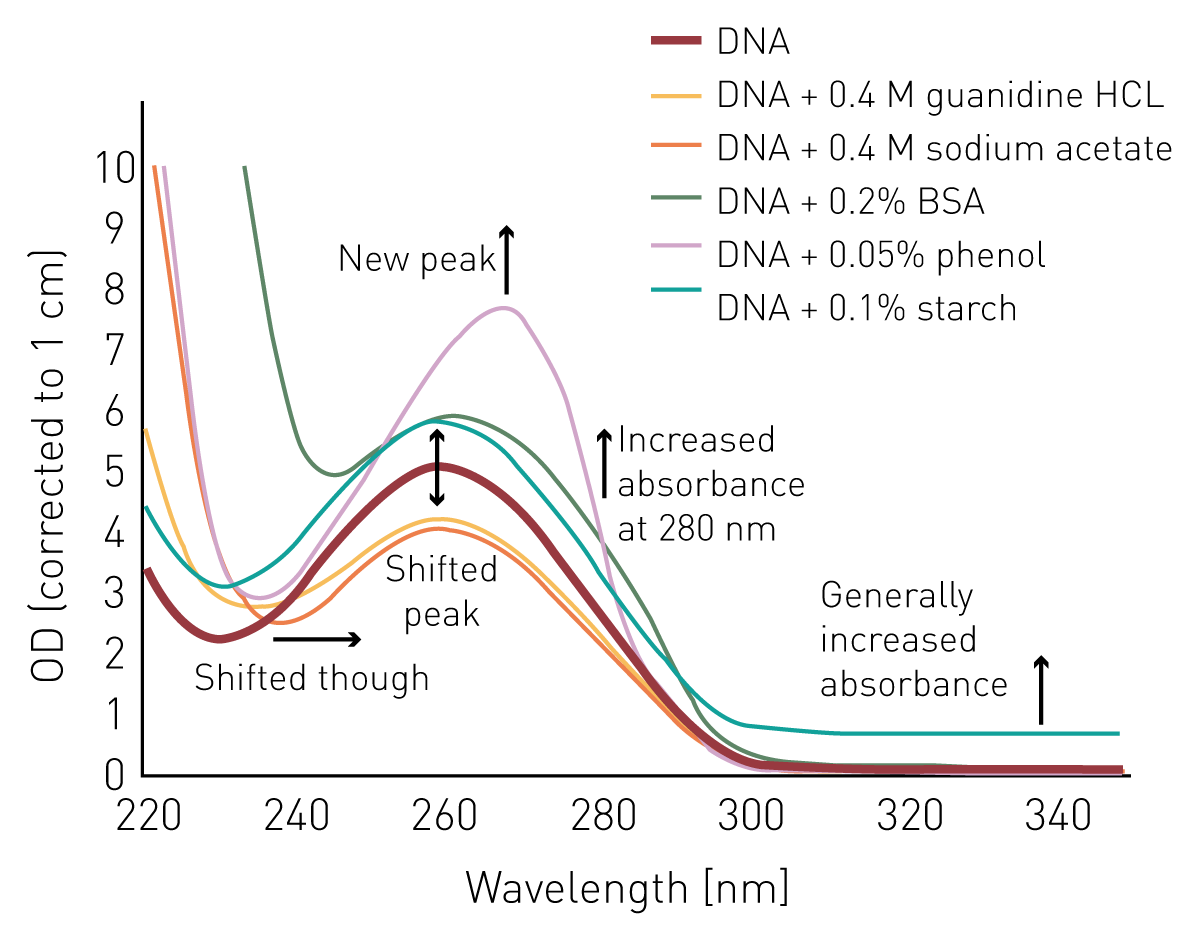Supreme Tips About How To Check Dna Concentration

Finger vortex your samples and spin down.
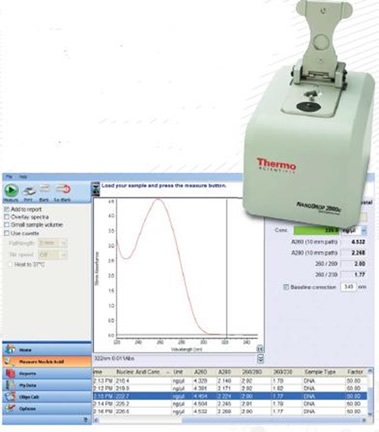
How to check dna concentration. Where, 100 is the dilution factor and 0.1 ml is the total volume of the dna. The concentration of dna can be estimated by running it on an agarose gel. Nanodrop reading for this blood sample after reverse transcription :1500ng/ul.
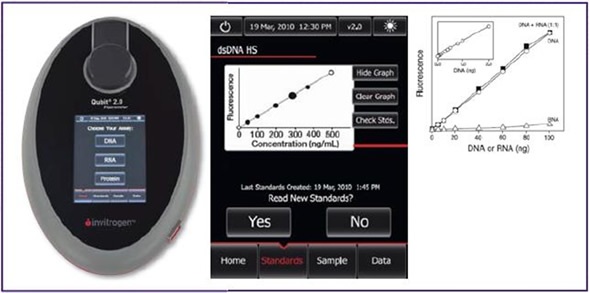
Spectrophotometric measurement of dna concentration. After amplifying my target gene (1900bp) into the required cdna by pcr,>cut specific gel bands and purify the gel and the concentration was 30ng/ul. Dna concentration is usually determined by the ultraviolet (uv) absorption, fluorescence staining, and diphenylamine reaction methods.
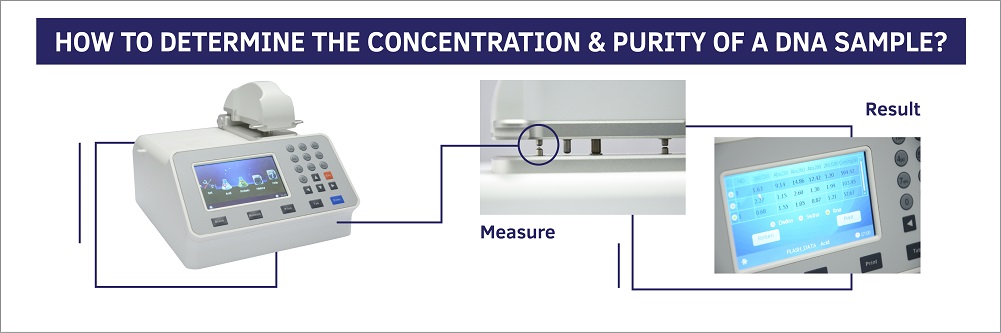
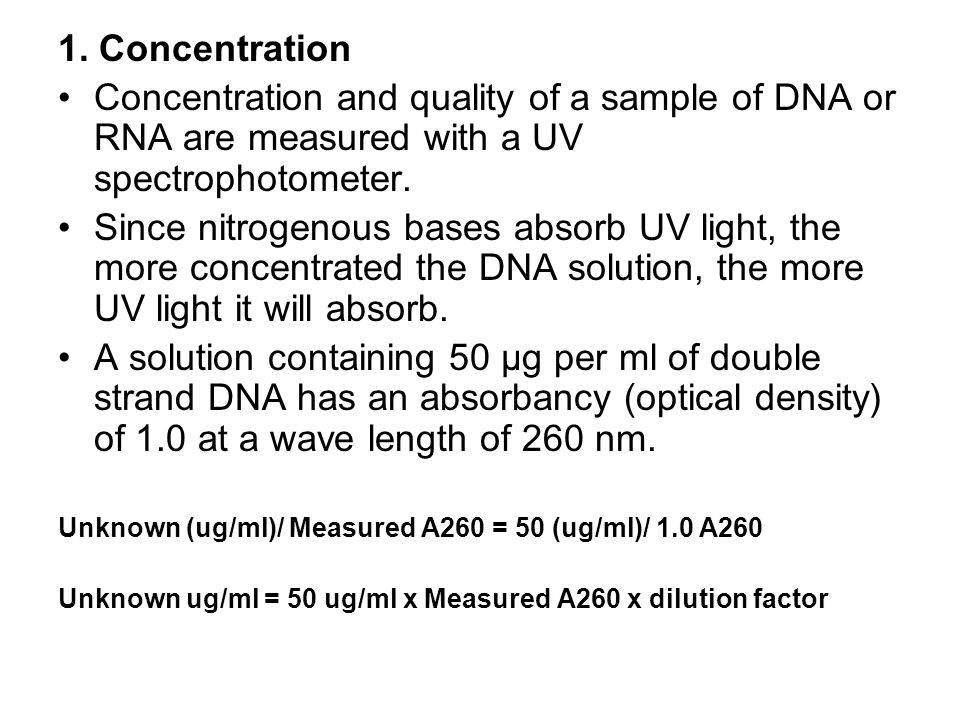
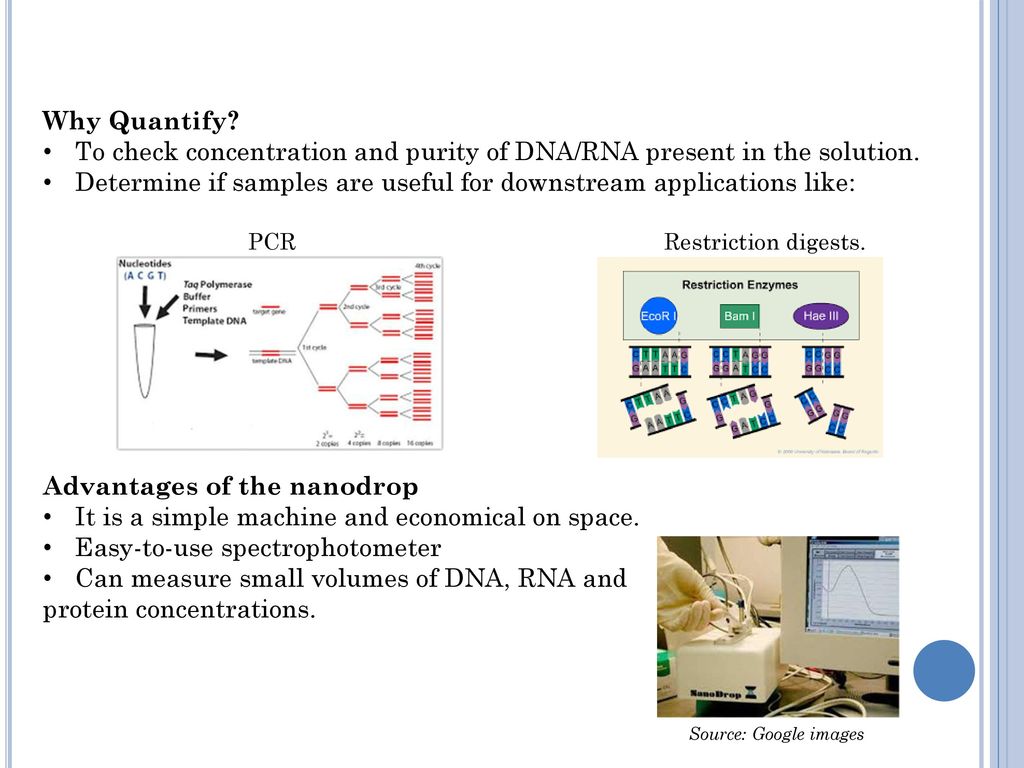
Measuring dna concentration by nanodrop objective to quantify the amount of dna in a phage or genomic dna sample. Here is the way to calculate dna concentration: Dna concentration is estimated by measuring the absorbance at 260nm, adjusting the a 260 measurement for turbidity (measured by absorbance at 320nm),.
This is especially common when researchers use. Fluoresces orange in uv light, making it possible to visualize the dna (figure 3). Start the nanodrop laptop and ensure the usb cable to the nanodrop is connected.
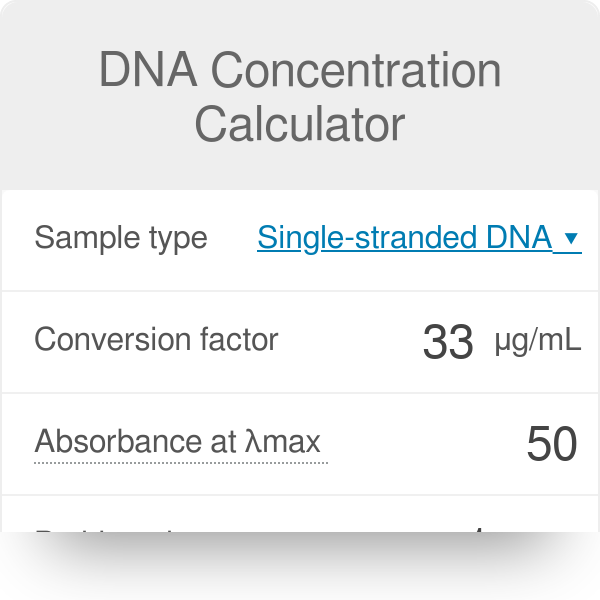
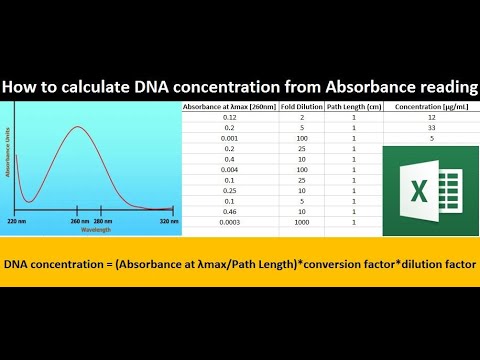
1 total dna (ug) = (a260) (50 ug/ml/a 260) (100) (0.1 ml) 2. However, the best method for. Dna concentration can be determined by measuring the absorbance at 260 nm (a 260) in a spectrophotometer using a quartz.
Ethanol precipitation is a popular method for desalting and concentrating dna. Close the lid and click measure, be sure to record the concentration and purity. It is best to dilute the.